| ON THE COVER | 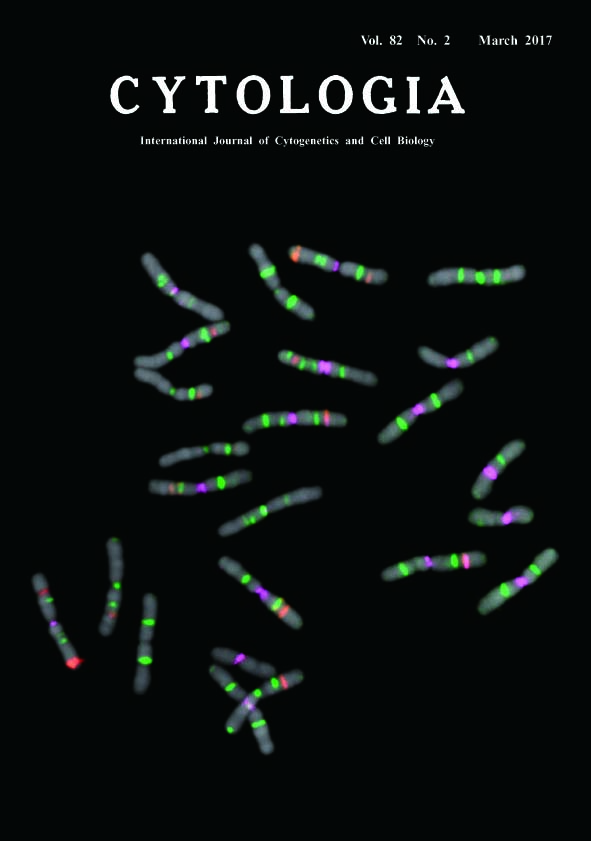 |
|
|---|---|---|
| Vol. 82 No.2 March 2017 | ||
| Invited Technical Note | ||
|
|
||
Fluorescence In Situ Hybridization on Chromosome Spread of Pinus tabulaeformis* Fukashi Shibata**, Yukari Matsusaki and Masahiro Hizume Faculty of Education, Ehime University, 3 Bunkyo, Matsuyama, Ehime 790–8577, Japan Received February 5, 2017; accepted February 28, 2017 In genus Pinus, fluorescence in situ hybridization (FISH) is a powerful tool for karyotyping. Each species has unique FISH signal patterns, and the difference of patterns reflect their phylogenetical relationships (Shibata et al. 2016). The seeds of P. tabulaeformis were sown on sterilised filter papers in petri dishes. After 10–14 d, the primary root tips were excised and treated with 0.05% colchicine for 12 h at 20°C, and fixed in a chilled solution of ethanol : chloroform : acetic acid (2 : 1 : 1) overnight. Fixed root tips were macerated in an enzyme mixture containing 2 or 3% cellulase Onozuka RS (Yakult), 0.5% pectolyase Y23 (Seishin), and 5 mM EDTA in 2×SSC buffer (pH 4.5) at 37°C for 30–60 min. The meristematic cells were squashed under coverslips on glass slides. The coverslips were removed by the dry-ice method; the glass slides were then air dried and stored in a freezer. The Arabidopsis-type telomere sequence repeats (TTT AGG G)n were PCR amplified (Ijdo et al. 1991) and labelled with biotin using the BioNick Labeling System (Invitrogen), and the signals were visualized with streptavidin-Cy5 (Invitrogen) and colored green on a pseudocolour image. We used EcoRI-digested plasmid with inserted wheat 35S rDNA (pTa71; Gerlach and Bedbrook 1979) and 5S rDNA amplified by PCR (Hizume 1993) from Rumex acetosa genomic DNA. We labelled the 35S and 5S rDNAs with digoxigenin (DIG) using DIG-High Prime (Roche); these signals were visualized with anti-DIG rhodamine conjugate (Roche) and shown as red on the pseudocolour image. The PCSR was a Pinus specific repetitive sequence (Hizume et al. 2001), amplified from the plasmid DNA of PDCD501 and then labelled with reactive isothiocyanate fluorescein (FITC) using FITC-High Prime (Roche); signals were colored magenta on pseudocolour image. Our FISH procedure followed the methodology of Hizume et al. (2002) and Shibata et al. (2016). The slides were counterstained with DAPI. We recorded the FISH signals with a cooled CCD camera (Sensys 1400, Photometrics), and pseudocolour images were generated using IPLab (Scanalytics).
Gerlach, W. L. and Bedbrook, J. R. 1979. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 7: 1869–1885. Hizume, M. 1993. Chromosomal localization of 5S rRNA genes in Vicia faba and Crepis capillaris. Cytologia 58: 417–421. Hizume, M., Shibata, F., Maruyama, Y. and Kondo, T. 2001. Cloning of DNA sequences localized on proximal fluorescent chromosome bands by microdissection in Pinus densiflora Sieb. & Zucc. Chromosoma 110: 345–351. Hizume, M., Shibata, F., Matsusaki, Y. and Garajova, Z. 2002. Chromosome identification and comparative karyotypic analyses of four Pinus species. Theor. Appl. Genet. 105: 491–497. Ijdo, J. W., Wells, R. A., Baldini, A. and Reeders, S. T. 1991. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 19: 4780. Shibata, F., Matsusaki, Y. and Hizume, M. 2016. A comparative analysis of multi-probe fluorescence in situ hybridisation (FISH) karyotypes in 26 Pinus species (Pinaceae). Cytologia 81: 409–421. *Corresponding author, e-mail: shibatafukashi@hotmail.com DOI: 10.1508/cytologia. 82.99 |
||