| ON THE COVER | 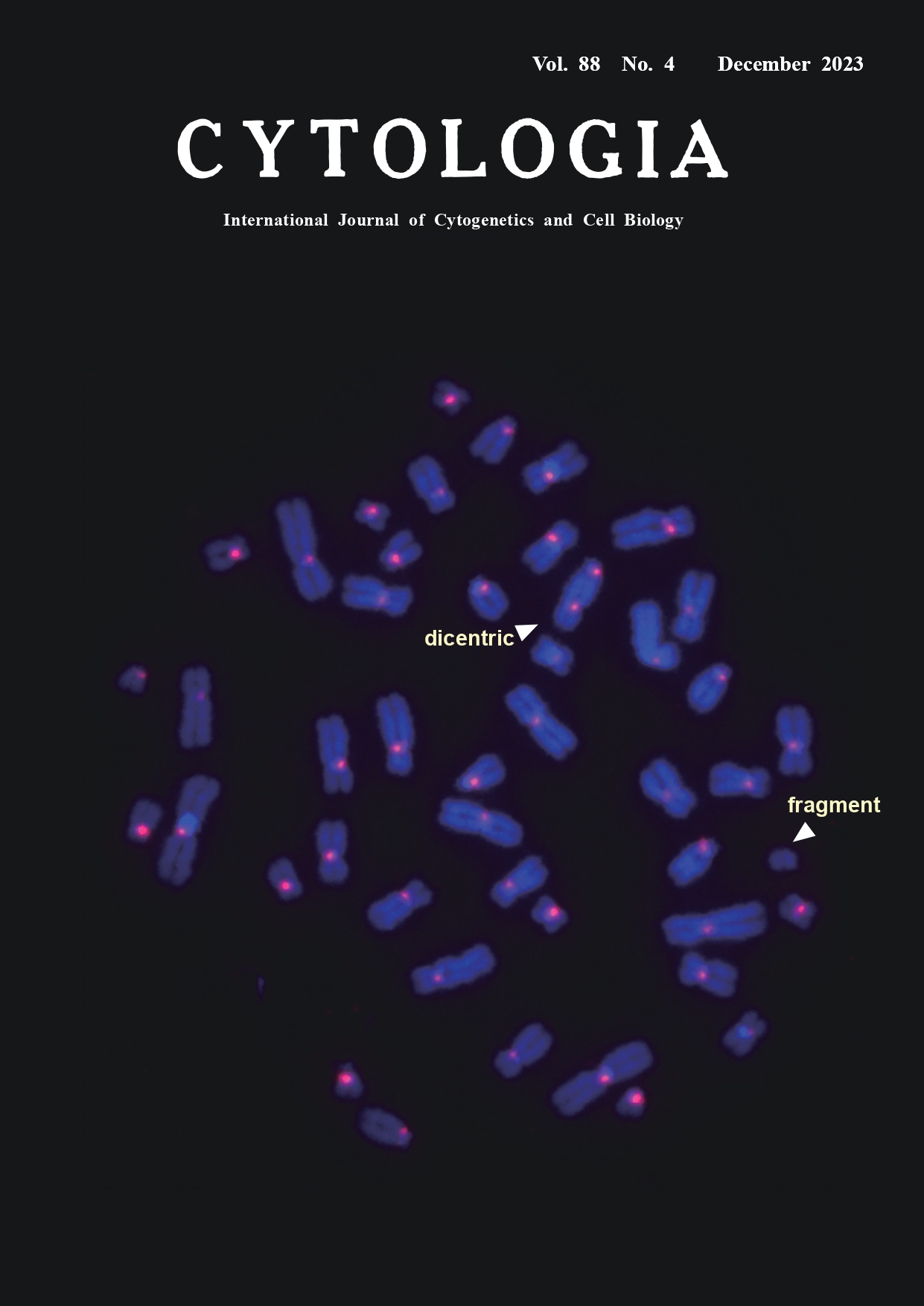 |
||
|---|---|---|---|
| Vol. 88 No.4 December 2023 | |||
| Technical Note | |||
|
|
|||
Application of automated scoring system of dicentric chromosome for biodosimetry Kotaro Ishii1, Miho Akiyama1, Kenji Kamimoto2, Hiroki Kawai2, and Yumiko Suto1* 1National Institutes for Quantum Science and Technology, 4–9–1 Anagawa, Inage-ku, Chiba 263–8555, Japan 2LPIXEL Inc., 1–6–1 Otemachi, Chiyoda-ku, Tokyo 100–0004, Japan
In cases of mass-casualty radiation accidents/incidents, a rapid dose assessment is required for patient triage and medical management decision. Biodosimetry is a method of estimating the exposure doses of patients using chromosomal aberrations as biological markers. The yield of dicentric chromosomes has a low background level and increases with radiation dose, and it is considered the most reliable and specific indicator of recent acute exposure to ionizing radiation. In dicentric chromosome assay (DCA), the number of dicentrics per peripheral blood lymphocyte of a radiation-exposed individual is applied to a dose–response curve that has been established by in vitro exposure experiments in advance. In conventional DCA, the number of dicentrics is scored in microscopy images of Giemsa-stained metaphases. Although centromeric regions of dicentrics are morphologically detectable as primary constrictions, they are sometimes unclear and difficult to distinguish from those in twisted chromosomes. The cover figure shows a typical metaphase image acquired by the fluorescence in situ hybridization technique with a peptide nucleic acid probe for centromeric alphoid DNA (PNA-FISH) as previously described (Suto et al. 2012) with some modifications. In brief, human peripheral blood samples were exposed to 3.0 Gy gamma-rays in vitro and cultured for 48 h. Chromosomal preparations were denatured in an alkaline solution of 0.1 M NaOH/ ethanol (7 : 3) at room temperature for 1 min and dehydrated in ethanol series. A pan-centromere PNA probe (F3003; Panagene, South Korea) was used. A total of 60 μL of a probe mix (60% formamide in 2×SSC, 10 ng of salmon sperm DNA, 5 ng of the PNA probe) was denatured at 90°C for 5 min, dropped onto a slide, covered with a piece of plastic sheet and kept at room temperature for 1 h in the dark. Then, the slide was washed in a posthybridization solution (2×SSC/0.1% Tween-20) for 10 min at 57°C, air-dried, and counterstained with 125 ng mL-1 4,6-diamidine-2-phenylindole (DAPI). Metaphase images were automatically captured with a cytogenetic scanning system (Metafer 4, Metasystem, Germany). Centromeric regions can be clearly recognized as red signals, which enable the differentiation between a dicentric chromosome (two signals) and a twisted chromatid chromosome (single signal). As the visual scoring of dicentrics is time-consuming and requires skill (100 to 500 min per 100-image scoring by an expert), efforts have been made to develop an automated scoring system. We assessed the performance of a commercially available automatic scoring software system (DCScore, Metasystem, based on machine learning algorithm) for detecting dicentrics in Giemsa-stained metaphase images. In testing 1,751 images (approximately 80,000 chromosomes), 57% of all chromosomes were identified as “chromosome”. Among all “dicentric chromosomes” identified by DCScore, the rate of true dicentrics confirmed by an expert was 10% (true positive). We have been developing a new scoring software system for detecting dicentrics in PNA-FISH metaphase images. Our in-house software employs deep learning algorithms. We evaluated a pilot version of the software using 1,000 PNA-FISH metaphase images as training and validation data. Then, 177 metaphase images from 3.0 Gy-irradiated cells were tested. The rate of correct identification of “chromosome” was as high as 98%. Although there remains the technical issue concerning the lower true positive rate (53%) in the software, it should be noted that the time needed was less than 1 min per 100-image scoring, indicating its promising usefulness in patient triage in large-scale radiation accidents/incidents. To solve the technical issue, we are still improving the system through intensive training using a much larger number of metaphase images.
Suto, Y., Hirai, M., Akiyama, M., Suzuki, T., and Sugiura, T. 2012. Sensitive and rapid detection of centromeric alphoid DNA in human metaphase chromosomes by PNA fluorescence in situ hybridization and its application to biological radiation dosimetry. Cytologia 77: 261–267. * Corresponding author, e-mail: suto.yumiko@qst.go.jp DOI: 10.1508/cytologia.88.281 |
|||