| ON THE COVER | 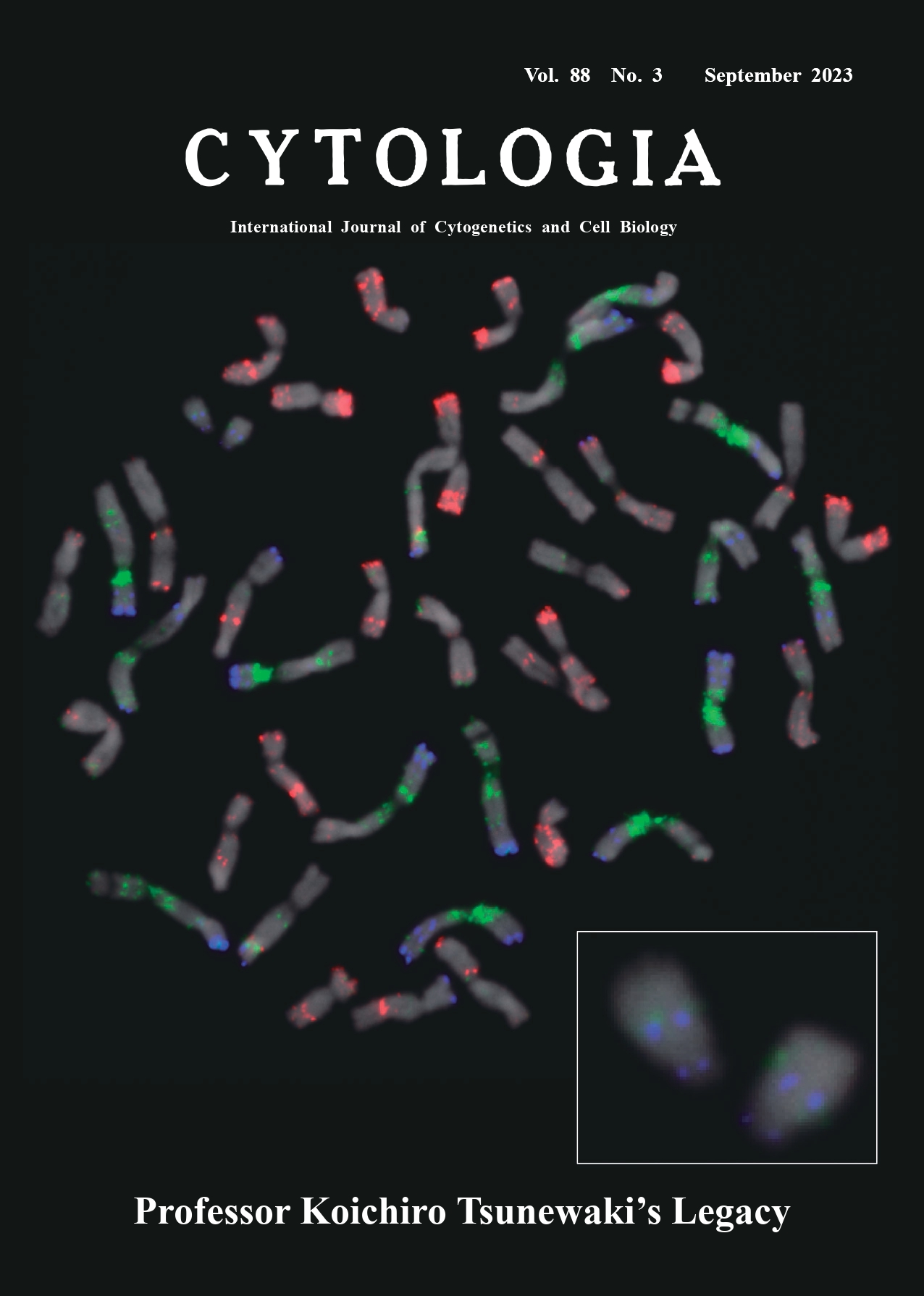 |
||
|---|---|---|---|
| Vol. 88 No.3 September 2023 | |||
| Technical Note | |||
|
|
|||
Molecular cytological identification of the gametocidal chromosome discovered by Endo and Tsunewaki (1975) in an alloplasmic wheat Shuhei Nasuda1*, Kazuki Murata1, Hiroyuki Kakui1,2, Koichi Yamamori1 and Takanori Yoshikawa1,3 1Graduate School of Agriculture, Kyoto University, Kyoto, Kyoto 606–8502, Japan 2Graduate School of Agricultural and Life Science, The University of Tokyo, Nishitokyo, Tokyo 188–0002, Japan 3National Institute of Genetics, Mishima, Shizuoka 411–8540, Japan
In this issue of Cytologia, a series of manuscripts are dedicated to the late Professor Koichiro Tsunewaki (1930–2022), who has been the mentor of wheat researchers worldwide. We often recognize him the researcher who developed the series of alloplasmic wheat lines that provided great opportunities to study maternal lineage of evolution of Triticum and Aegilops species, to elucidate interactions of nuclei and cytoplasms, and to improve wheat by hybrid wheat breeding. Eventually, his research interest was broader. He found the gametocidal (Gc) genes, nuclear genes causing hybrid sterility, in the backcrossing generations of alloplasmic wheat. Following the first report of the Gc gene by Endo and Tsunewaki (1975), we now know at least five different Gc genes in wheat. Here, we demonstrate spontaneously modified chromosomes of the first found Gc chromosome 3Ct from Aegilops triuncialis L. (2n=4x=28, genome constitution CtCtUtUt) in a wheat (Triticum aestivum L., 2n=6x=42, AABBDD) cultivar Chinese Spring. Endo (2007) reported a spontaneous structural rearrangement of chromosome 3Ct , namely del(3CL), that is a telosome with terminal deletion of the long arm of 3Ct (enlarged in the inset at the bottom right of the cover). The del(3CL) chromosome maintains the Gc function, which indicates that the Gc3-C1 gene locates in the peric entromeric region of chromosome arm 3CtL. Currently, the disomic addition of del(3CL) is available with the accession number LPGKU2162 from NBRP-Wheat, Japan (https:// shigen.nig.ac.jp/wheat/komugi/). The cover figure shows a metaphase spread of a cell in the root tip meristem as probed with three repetitive element probes. Wheat has been a good material for cytogenetics because of its large chromosomes size and therefore different molecular-cytogenetic techniques have been adapted. Non-denaturing fluorescence in situ hybridization (ND-FISH) technology of localized repetitive elements is less labor and time intensive than conventional FISH and, more importantly, provide higher signal/noise ratio when adapted to wheat chromosomes. We applied three probes: Oligo-pSc119.2-1 (TAMRA- 5′-CCG TT TTG TG GAC TA TTA CT CAC CG CTT TG GGG TC CCA TA GCT AT-3′) detecting mainly the Bgenome chromosomes (pseudo-colored in blue), OligopTa535- 2 (Alexa Fluor 488-5′-GAC GA GAA CT CAT CT GTT AC ATG GG CAC TT CAA TG TTT TT TAA AC TTA TT TGA AC TCC A-3′) localized mainly on the D-genome chromosomes (red), and Oligo-CTT6 (Alexa Fluor 647-5′-CTT CT TCT TC TTC TT CTT-3′) to distinguish C genome chromosomes (green) (Mirzaghaderi et al. 2014, Tang et al. 2014). The Gc chromosome del(3CL) carries diagnostic foci of Oligo-pSc119.2-1 and Oligo-CTT6 in the interstitial region of the truncated long arm (shown in the inset of the cover). The smaller size of the Gc chromosome del(3CL) than normal meta- or submeta-centric wheat chromosomes make us possible to flow-sort them from others and obtain shotgun sequences of the target chromosome (Murata et al. unpublished), which will allow us to identify the Gc3-Ct1 gene on del(3CL) through molecular genomic analyses.
Endo, T. R. 2007. The gametocidal chromosome as a tool for chromosome manipulation in wheat. Chromosome Res. 15: 67–75. Endo, T. R. and Tsunewaki, K. 1975. Sterility of common wheat with Aegilops triuncialis cyoplasm. J. Hered. 66: 13–18. Mirzaghaderi, G., Houben, A. and Badaeva, E. D. 2014. Molecularcytogenetic analysis of Aegilops triuncialis and identification of its chromosomes in the background of wheat. Mol. Cytogenet. 7: 91. Tang, Z., Yang, Z. and Fu, S. 2014. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55: 313–318. * Corresponding author, e-mail: nasuda.shuhei.5z@kyoto-u.ac.jp DOI: 10.1508/cytologia.88.167 |
|||