| ON THE COVER | 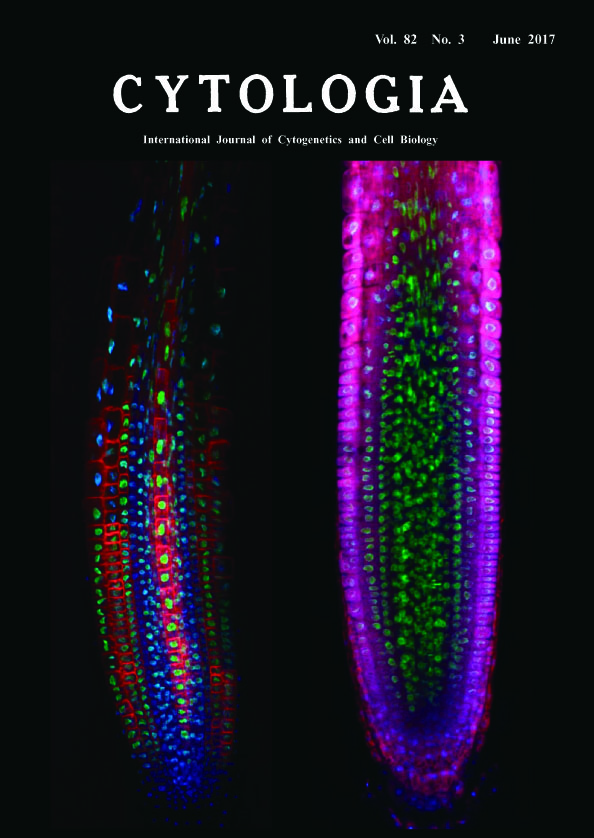 |
|
|---|---|---|
| Vol. 82 No.3 June 2017 | ||
| Technical Note | ||
|
|
||
Deep Imaging of Plant Roots by a Rapid Transparency Technique TOMEI Yuki Sakamoto1 and Sachihiro Matsunaga1,2* 1 Imaging Frontier Center, Research Institute for Science and Technology, Tokyo University of Science, 2641 Yamazaki, Noda, Chiba 278–8510, Japan 2 Department of Applied Biological Science, Faculty of Science and Technology, Tokyo University of Science, 2641 Yamazaki, Noda, Chiba 278–8510, Japan Received February 6, 2017; accepted May 31, 2017 Fluorescent signals from a depth of several hundred micrometers or more in organs and tissues are decreased by reflection, scattering and absorption of light. To resolve the problem in microscopic observation, we have developed a novel and rapid clearing method TOMEI (Transparent plant Organ MEthod for Imaging) (Katagiri et al. 2016, Hasegawa et al. 2016). We could prepare transparent organs of Arabidopsis thaliana and leaves of Oryza sativa for three to six hours without quenching of fluorescent proteins (Hasegawa et al. 2016). We show here the comparison of longitudinally optical sections between a transparent root by TOMEI (left image) and an only-fixed root in PBS (right image). Roots of seven-day-old A. thaliana seedlings expressing histone H2B (H2B)-GFP and low-temperature and salt responsive protein 6b (LTI6b)-tdTomato were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 60 min at 25°C and then incubated with 5 μg mL-1 4',6-diamidino-2-phenylindole (DAPI) in PBS for 60 min at 25°C to stain nuclei. Specimens were treated with 20% TOMEI solution (http://www.tcichemicals. com/eshop/ja/jp/commodity/T3530/, Tokyo Chemical Industry, Tokyo, Japan) for 10 min at 25°C. Finally, TOMEI treated specimens were mounted with 20% TOMEI solution while fixed specimens were mounted with PBS. These specimens were observed by a confocal scanning laser microscope FV1200 (Olympus, Tokyo, Japan), equipped with ×40 objective (N.A. 1.3, Olympus). DAPI, GFP, and tdTomato were exited with 405-nm, 473-nm, and 559-nm diode lasers, respectively. Optical section images at the center of root were processed with ImageJ 1.49v (National Institute of Health, Bethesda, MD, USA). Blue, green and red represents DAPI, GFP and tdTomato fluorescence, respectively. GFP signals have the most strong and stable intensity of these fluorescence because there is no difference in the fluorescence pattern from the inner tissues between in TOMEI-treated and only-fixed roots. Although both H2B-GFP and DAPI fluorescence were derived from nuclei, DAPI signals in the only-fixed root could be mainly detected from epidermal and cortex cell layers, whereas those in TOMEI-treated root could be detected from all cell layers including the most inner tissue, stele. LTI6btdTomato signals in the stele could be also detected only after TOMEI treatment. The reason why both signals of DAPI and LTI6b-tdTomato were acquired from the inner tissues in TOMEI-treated root can be explained by the reduction of background signals from different focus planes. TOMEI is an excellent technique to inhibit scattering and reflection of light in plant organs. We believe that TOMEI will contribute to reveal the morphology of cells, organelles, and tissues and the localization pattern of proteins, DNA, RNA and some metabolites in the inside of multi-cellular organisms.
Hasegawa, J., Sakamoto, Y., Nakagami, S., Aida, M., Sawa, S. and Matsunaga, S. 2016. Three-dimensional imaging of plant organs using a simple and rapid transparency technique. Plant Cell Physiol. 57: 462–472. Katagiri, Y., Hasegawa, J., Fujikura, U., Hoshino, R., Matsunaga, S. and Tsukaya, H. 2016. The coordination of ploidy and cell size differs between cell layers in leaves. Development 143: 1120–1125. *Corresponding author, e-mail: sachi@rs.tus.ac.jp DOI:10.1508/cytologia.82.221 |
||