Our Research
Autophagy
While all components of our bodies are continually synthesized, they are also constantly eliminated. Whole organisms and even individual cells are maintained in optimal condition through the recycling of their own constituents, which also allows them to adapt to various internal and external changes.
Autophagy
Autophagy is a cellular process by which cytoplasmic materials, including proteins and organelles, are delivered to and degraded in lysosomes. Autophagy has multiple functions such as basal turnover of cellular constituents, elimination of unwanted materials, and recycling of nutrients for self-nourishment. As many molecules involved in autophagy have been identified, we can now study autophagy at the molecular level. In fact, autophagy has become one of the hottest topics in life science.
Our Research
In our laboratory, we study the mechanisms and physiological functions of autophagy and other degradation processes, specifically:
- molecular mechanisms of autophagosome formation and maturation, and autophagosome-lysosome fusion
- physiological and pathophysiological roles of autophagy using mice and zebrafish
- development of autophagy-monitoring methods
- basic research on autophagy-related diseases
- mechanisms of intracellular degradation of membranes
- dynamics of organelles
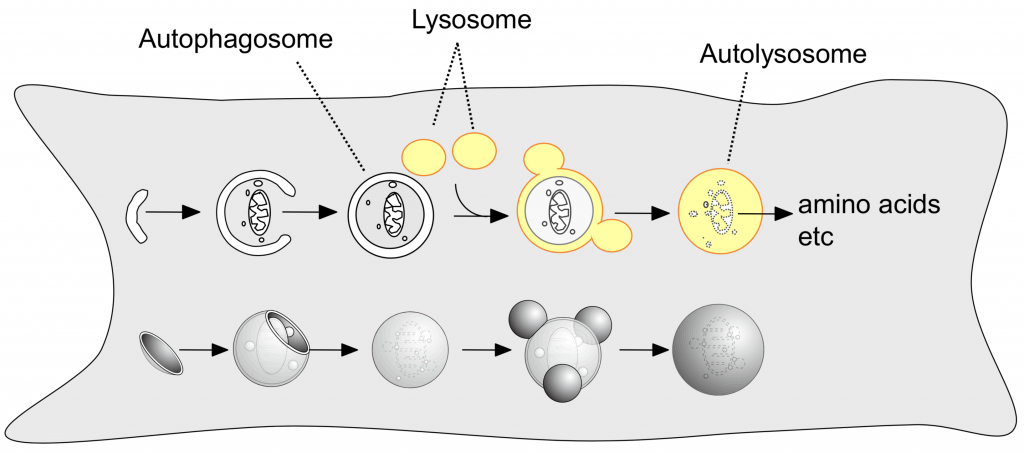
A small membrane cisterna (called the isolation membrane or phagophore) elongates to enclose
a part of the cytoplasm and become the autophagosome. Lysosomes fuse with the outer auto-
phagosomal membrane to degrade the inner autophagosomal membrane and enclosed contents.

