| ON THE COVER | 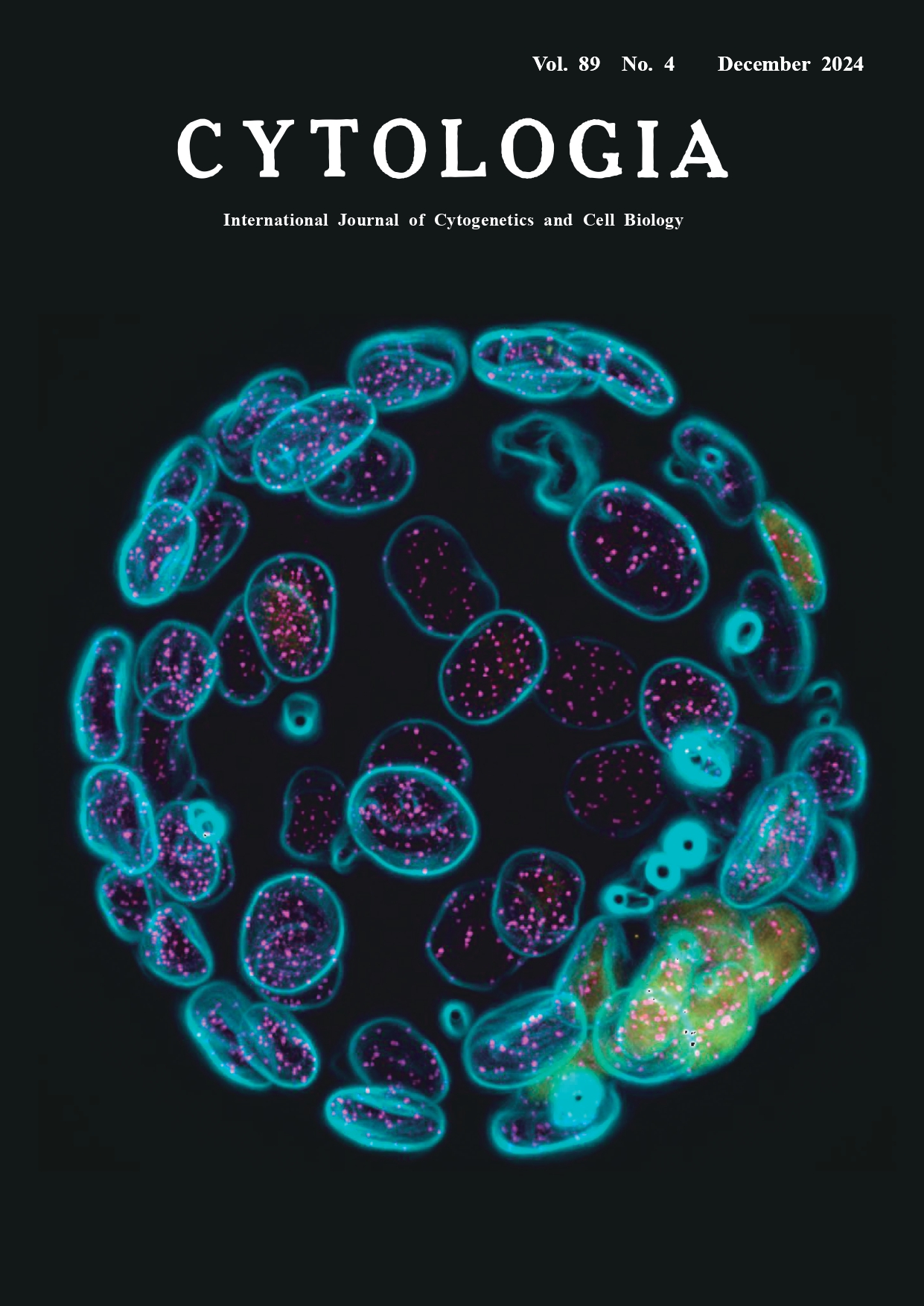 |
||
|---|---|---|---|
| Vol. 89 No.4 December 2024 | |||
| Technical Note | |||
|
|
|||
| Development of a microscopy-based method for genome ploidy analysis of
all constituent cells in mouse blastocysts Koya Shimabukuro1 and Miho Ohsugi1,2* 1 Department of Life Sciences, Graduate School of Arts and Sciences, The University of Tokyo, 3–8–1 Komaba, Meguro-ku, Tokyo 158–8902, Japan 2 Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 7–3–1 Hongo, Bunkyo-ku, Tokyo 113–8654, Japan
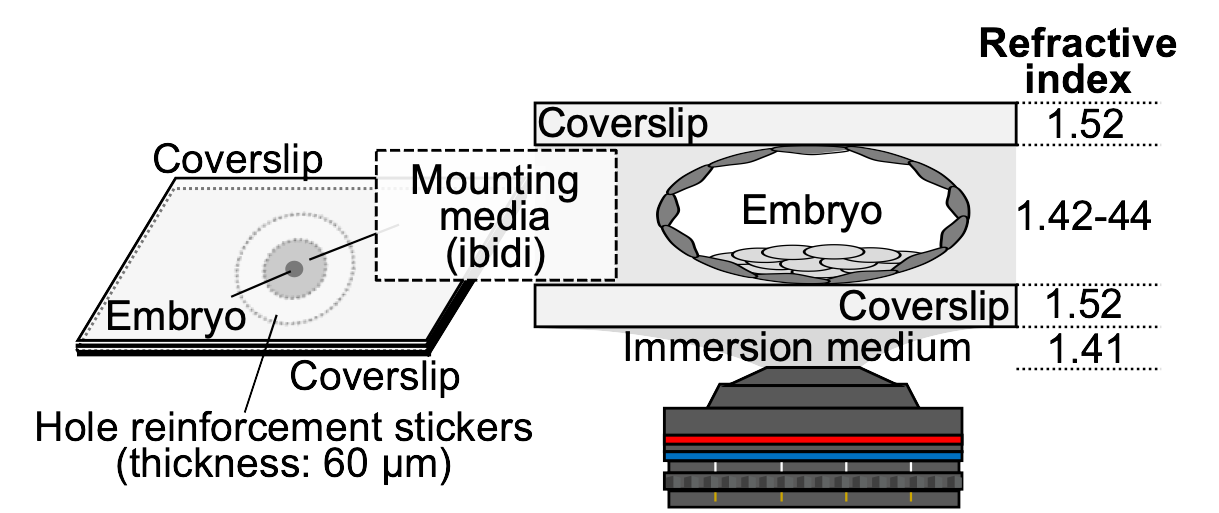
Embryonic stem cells established from mouse haploid embryos usually contain diploid cells, but when and how genome doubling occurs remains poorly understood (Liu and Li 2022). To examine whether genome doubling occurs during preimplantation development, we established a procedure to verify the ploidy of cells constituting blastocysts derived from parthenogenetic mouse eggs. Ploidy was assessed by immunostaining blastocysts with an anti-CENP-A antibody, a centromere marker, and counting the number of CENP-A signals in the nuclei. Imaging the entire blastocyst, which is approximately 100 μm thick, with sufficient quality to count CENP-A signals is challenging because spherical aberration (blurring caused by the lens) increases with the numerical aperture of the lens and the distance from the coverslip. When using an oil-immersion objective lens on aqueous samples, the observation depth is restricted to about 20 μm from the coverslip. To overcome this difficulty, we developed a method for preparing samples for observation by sandwiching the blastocyst between two coverslips with a 60-μm thick spacer (Fig. 1). We also matched the refractive indices of the mounting media and the lens immersion medium. Using this method, the blastocyst can be mildly flattened to 60–70 μm in thickness for microscopy, and by flipping the sample, it is also possible to observe the embryo from the opposite side. Combining observation data from both sides allows for high-resolution imaging of the entire embryo. The cover figure shows an immunofluorescent image of a mouse diploid blastocyst (CENP-A: magenta, Oct3/4: yellow, Hoechst: cyan). Immunofluorescence staining was performed according to the previously reported method (Totsuka and Ohsugi 2020), with the following modifications for staining with the anti-CENP-A antibody: after permeabilization, samples were treated with Lambda protein phosphatase (P0753S, New England Biolabs). The antibodies and DNA dye used are as follows: CENP-A (2048, Cell Signaling Technology), Oct3/4 (c-5279, Santa Cruz Biotechnology), and Hoechst 33342 (H-1399, Thermo Fisher Scientific). After immunostaining, the embryos were gradually replaced in 25%, 50%, 75%, and 100% ibidi mounting medium (50001, ibidi) and finally placed inside hole reinforcement stickers (ML-251, Nichiban) and sandwiched between two coverslips. Z-stack images were collected at 0.5-μm intervals (140 optical z-stacks) using a fluorescence microscope (IX70, OLYMPUS) equipped with a 60×/1.30 NA silicone immersion objective lens (OLYMPUS) and a cooled CCD camera (CoolSNAP HQ, Roper Scientific), controlled by DeltaVision SoftWorx software (Applied Precision). The Hoechst channel was processed using the ‘Find Edges’ function in Fiji in order to enhance the visibility of CENP-A signals in the merged images. Background subtraction was also applied to the CENP-A channel. The three channels (Hoechst, CENP-A, and Oct3/4) were then merged, and all 140 z-stacks were processed using maximum intensity projection to generate the final image. This method allows us to determine the ploidy of all cells constituting the blastocyst.
Fig. 1. Observation method and refractive index.
Totsuka, T. and Ohsugi, M. 2020. Production of mouse androgenetic embryos using spindle perturbation. Sci. Rep. 10: 6556. * Corresponding author, e-mail: mihoohsugi@g.ecc.u-tokyo.ac.jp DOI: 10.1508/cytologia.89.261 |
|||