| ON THE COVER | 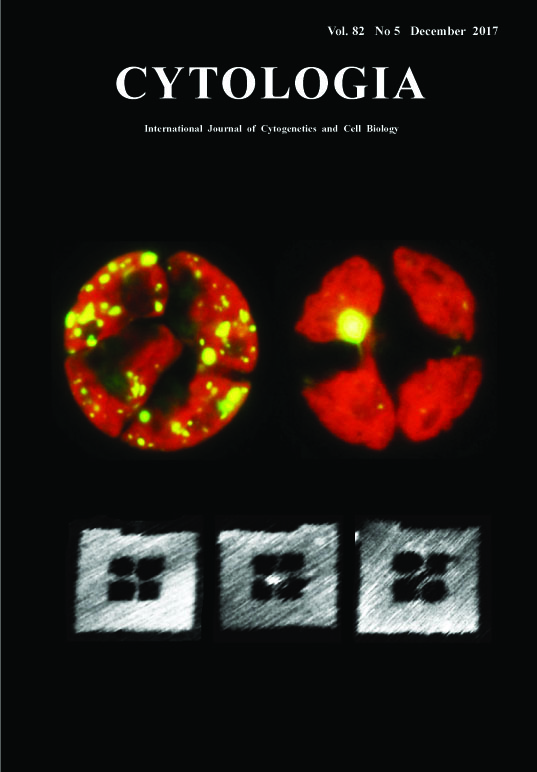 |
|
|---|---|---|
| Vol. 82 No.5 December 2017 | ||
| Technical Note | ||
|
|
||
Finding Holliday Junction Resolvases: A Crucial Factor for Chloroplast Nucleoid Segregation Yusuke Kobayashi1, Osami Misumi2 and Yoshiki Nishimura1* 1 Laboratory of Plant Molecular Genetics, Department of Botany, Kyoto University, Oiwake-cho, Kita-Shirakawa, Kyoto 606–8502, Japan 2 Department of Biological Science and Chemistry, Faculty of Science, Graduate School of Sciences and Technology for Innovation, Yamaguchi University, Yamaguchi 753–8512, Japan Received September 17, 2017; accepted September 27, 2017 Chloroplasts generally possess multiple copies of chloroplast (cp) genomes which are organized into cpDNA–protein complexes, called cp nucleoids. Cp nucleoids change drastically during the chloroplast divisions, differentiation, life cycle and evolution. In the green alga Chlamydomonas reinhardtii, a vegetative cell has ca.80 copies of cpDNA packaged into ca.5 to 10 cp nucleoids. Upon chloroplast division, cp nucleoids become scattered throughout chloroplasts. To address the molecular mechanism of the cp nucleoid segregation, we analyzed the mutant monokaryotic chloroplast (moc) (Misumi et al. 1999), which possesses only a single cp nucleoid and shows unequal segregation (upper panel: unequal segregation of cp nucleoids are visualized by expressing YFP fused to a major chloroplast nucleoid protein, HU (yellow) in contrast with WT (left). Red is autofluorescence from chlorophyll). Eventually, a gene MOC1 encoding Holliday junction (HJ) resolvase was identified as a key factor that affects the dynamism of cp nucleoids (Kobayashi et al. 2017). HJ resolvase is an endonuclease that cuts the Holliday junction, a four-way DNA structure, in a sequence-structure specific manner. We employed a DNA origami technology in combination with high speed AFM (atomic force microscopy) (Suzuki et al. 2014) to visualize the action of MOC1 on HJs (lower left: before the reaction, middle: HJ bound by AtMOC1 (center), right: after the digestion). The DNA frame was assembled in a 20 μL solution containing 10 nM of M13mp18 single-stranded DNA (New England BioLabs, Ipswich, MA), 100 nM of staple strands (226 strands), 20 mM Tris–HCl (pH 7.6), 1 mM EDTA, and 10 mM MgCl2. The mixture was annealed from 85 to 15°C at a rate of -1.0°C/min. The pre-assembled Holliday junction structures were subsequently incorporated into the DNA frame by annealing the mixture from 40 to 15°C at a rate of -0.5°C/min. The sample was purified by gel-filtration chromatography. The reaction was carried out at room temperature for 10 min in solution containing DNA frame ca.10 nM, MBP-AtMOC1 ca.50 nM, 20 mM Tris–HCl (pH 7.6), 10 mM MgCl2, and 1 mM EDTA. After the reaction, 2 μL of the sample was deposited onto a freshly cleaved mica, and then rinsed with the same buffer. AFM imaging was performed using a high-speed AFM (Nano Live Vision, RIBM, Tsukuba, Japan). The sample was imaged in the buffer solution at room temperature with a silicon nitride cantilever (BLAC10EGS, Olympus Corporation, Tokyo, Japan) at a scan rate of 0.2 frames per second (fps).
Kobayashi, Y., Misumi, O., Odahara, M., Ishibashi, K., Hirono, M., Hidaka, K., Endo, M., Sugiyama, H., Iwasaki, H., Kuroiwa, T., Shikanai, T. and Nishimura, Y. 2017. Holliday junction resolvases mediate chloroplast nucleoid segregation. Science 356: 631–634. Misumi, O., Suzuki, L., Nishimura, Y., Sakai, A., Kawano, S., Kuroiwa, H. and Kuroiwa, T. 1999. Isolation and phenotypic characterization of Chlamydomonas reinhardtii mutants defective in chloroplast DNA segregation. Protoplasma 209: 273–282. Suzuki, Y., Endo, M., Cañas, C., Ayora, S., Alonso, J. C., Sugiyama, H. and Takeyasu, K. 2014. Direct analysis of Holliday junction resolving enzyme in a DNA origami nanostructure. Nucleic Acids Res. 42: 7421–7428. *Corresponding author, e-mail: yoshiki@pmg.bot.kyoto-u.ac.jp DOI:10.1508/cytologia.82.465 |
||