| ON THE COVER | 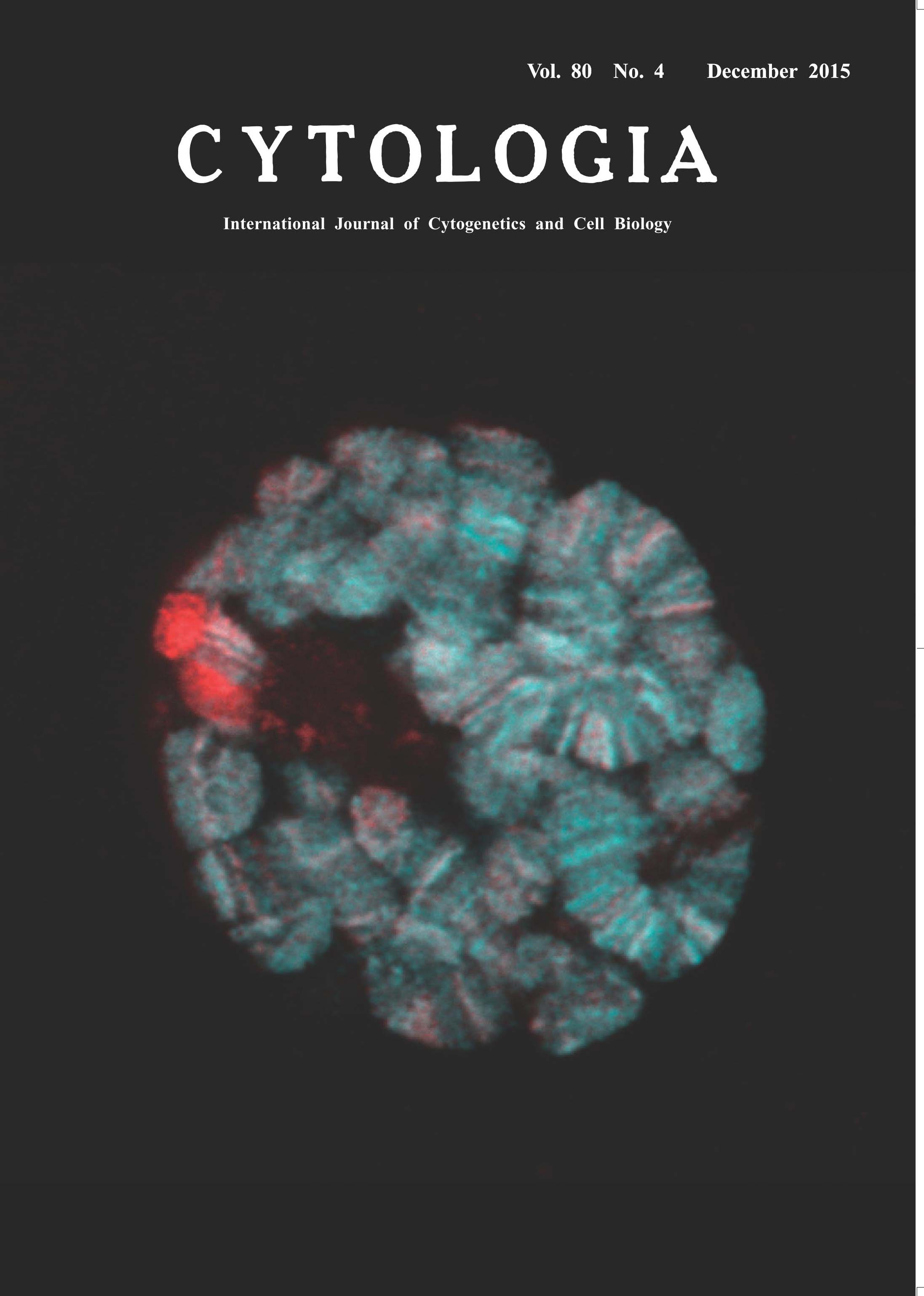 |
|
|---|---|---|
| Vol. 80 No.4 December 2015 | ||
| Technical note | ||
|
|
||
| Histone Acetylation on Drosophila Polytene Chromosomes Visualized by Mintbody Yuko Sato1, Masanori Mukai2and Hiroshi Kimura1* Post-translational modifications of histone proteins play an important role in gene expression and genome integrity. In response to extracellular stimuli, histone modifications change dynamically by modifying and demodifying enzymes. To visualize histone modifications in vivo, we developed a genetically encoded system using a fluorescent probe termed “mintbody” (modification-specific intracellular antibody). We cloned cDNA encoding variable regions of an antibody directed against histone H3 Lys9 acetylation to generate H3K9ac-mintbody by fusing the single chain variable fragment to enhanced green fluorescent protein (EGFP). H3K9ac-mintbodies were successfully expressed in cultured human cells, showing euchromatic pattern in living nuclei. H3K9ac-mintbody has enabled us to monitor the changes of acetylation levels in living cells in real time, as their nuclear concentration reflects the acetylation level. We generated transgenic fruit flies and zebrafish that express H3K9ac-mintbody, which retains high specificity to H3 Lys9 acetylation with minimal interference to cell function (Sato et al. 2013). The picture on the title page shows polytene chromosomes in the salivary gland of Drosophila melanogaster expressing H3K9ac-mintbody (Cyan); DNA is counterstained with Hoechst33342 (Red). The H3K9ac-mintbody coding sequence was cloned into the pUASp vector and the resulting plasmid was injected into y w flies to generate the transgenic flies carrying UAS-H3K9ac-mintbody. H3K9ac-mintbody was expressed in salivary gland cells under the control of hs-Gal4 driver. Salivary glands were isolated from larvae and placed onto a μ–dish (ibidi) in PBS. H3K9ac-mintbody allows live imaging of H3K9ac distribution on polytene chromosomes in individual cells. Here, to compare H3K9ac localization with DNA banding pattern, salivary glands were fixed and stained with Hoechst33342, and optical sections were collected using a confocal microscope (FV1000; Olympus). In polytene chromosomes, genes are organized into discrete bands. Transcriptionally- silent heterochromatin, or “chromocenter”, is densely stained with Hoechst33342 but devoid of H3K9ac-mintbody, consistent with the view that histone H3 acetylation marks potentially active chromatin. H3K9ac-mintbodies were concentrated in some bands more than the others, suggesting that H3K9 acetylation levels differ in individual genes. It would be interesting to see how the acetylation changes during gene activation in living cells using mintbodies.
1 Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, Yokohama, Kanagawa 226– 8501, Japan, 2Faculty of Science and Engineering and Graduate School of Natural Science, Konan University, Higashinada, Kobe, Hyogo 658–8501, Japan *Corresponding author, e-mail: hkimura@bio.titech.ac.jp DOI: 10.1508/cytologia.80.383 |
||