| ON THE COVER | 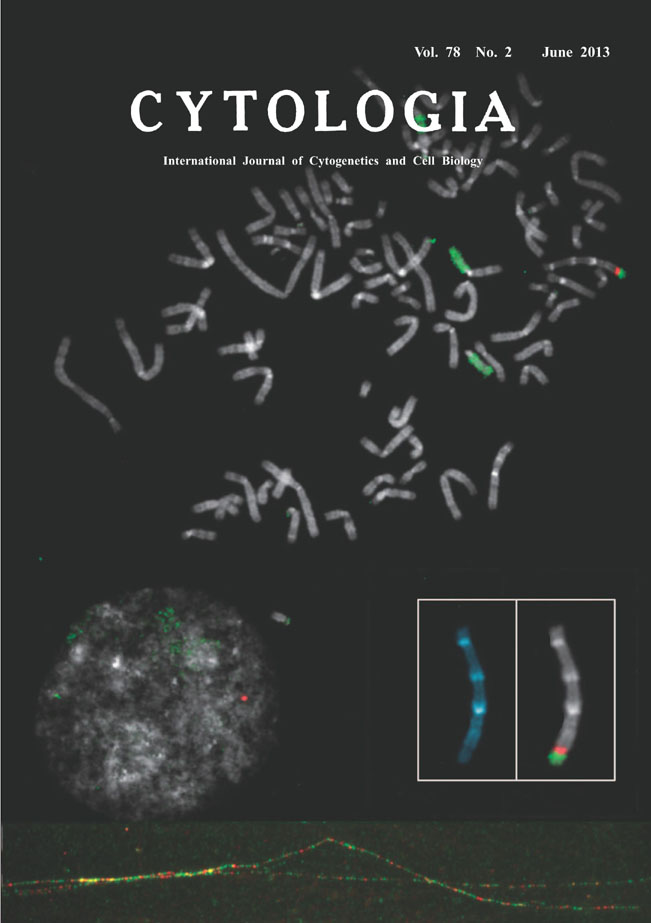 |
|---|---|
| Vol. 78 No.2 June 2013 | |
| Technical note | |
|
|
|
| Comparative Fiber-FISH Reveals What Happened at the Integration Site of the Transfected Plasmid DNA Transfection of the plasmid DNAs into somatic cells is one of widely used techniques in current molecular cell biology. Fluorescence intensity of stable transformants expressing proteins such as green fluorescent protein encoded by the transgene is usually monitored for selection of cell clones. Therefore, little is concerned with how they are integrated into the genome. The picture on the title page shows one metaphase and an interphase nuleus from human lung cancer cell line, H1299dA3-1 clone #1, that contained a single integration site of the transgene at the distal site of a subtelocentric chromosome (the chromosome is enlarged in white boxes, left; stained with DAPI and right; hybridized with DNA probes). Transgene of the pIRES-TK-GFP plasmid DNA (7,205 bp, see Ogiwara et al. 2011, Oncogene 30: 2135-2146) labeled with digoxigenin-11-dUTP was detected by anti- digoxigenin rhodamine (red) and counter-hybridization signals for FITC-painted chromosome #15 (green) were seen, simultaneously. To elucidate copy numbers of the transgene integrated in this site, we next applied stretched DNA fiber-FISH. Both of metaphases and extended DNA fibers were prepared on the same slide. To obtain extended DNA fibers, a droplet of the cell suspension was treated with lysis buffer containing 0.5% SDS, then the flowed contents from the cells were gently stretched by tilting the slides. The slides were air dried, fixed with Carnoy’s fixative, and processed for two-color FISH. The former plasmid DNA was divided into approximately two parts, a IRES-puro fragment (2.7 kb) and a CMV-TK-GFP fragment (2.7 kb). Each of the linearized fragments was labeled with non-radioactive haptens and visualized with FITC-avidin (green) or anti-digoxigenin rhodamine (red), respectively. As a result, stretched DNA (attached figure at the bottom) showed the single integration site actually contained multiple copies of IRES and GFP fragments spanning over hundred kilobases. In conclusion, numerous number of plasmid DNAs seems to be amplified at the target site when the transgene was incorporated into the genome possibly due to so-called concatamer formation by rolling cycle of replication. (Izuru Yokomi 1,3 , Hideaki Ogiwara 2 , Takashi Kohno 2 , Jun Yokota 2 , and Hitoshi Satoh 4 *, 1 Department of Pharmacology, School of Medicine, St. Marianna University, Kanagawa, 2 Biology Division, National Cancer Center Research Institute, Tukiji, Minato-ku, Tokyo, 3 Animal Care Co., Ltd., 4 Department of Medical Genome Science, Graduate School of Frontier Sciences, The University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639) |
|