| ON THE COVER | 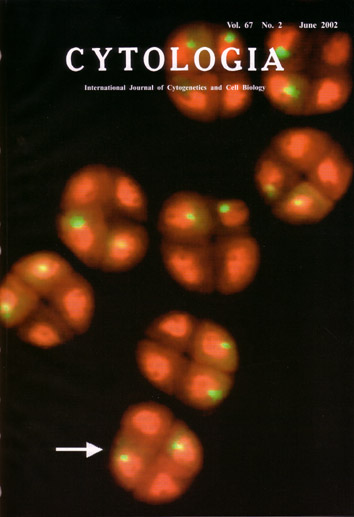 |
|---|---|
| Vol. 67 No.2 June 2002 | |
| Technical note | |
|
|
|
| Tetrad analysis is a genetical method to
locate genes and centromeres on a linkage map with a high degree of precision.
Despite its effectiveness and accuracy, application of this method has
been generally limited to fungi, algae and moss. Herein we demonstrate
a new method of tetrad analysis applicable to other organisms. This tetrad
analysis is combined with fluorescence in situ hybridization (FISH) and thus is called 'tetrad-FISH analysis'. We demonstrate
the effectiveness of this method using tetrads of rye. Secale cereal. The 'JNK' rye strain contains heterochromatin in the interstitial region of Chromosome 2R. We have previously cloned the tandem repetitive sequence forming this heterochromatin (pScJNK). We performed FISH using pScJNK as the probe to the tetrads of heterozygotes for the heterochromatin region. The frequency of tetrads demonstrating positive signals in the two cells arranged in the diagonal position (arrowed) must correspond to the recombination frequency between the heterochromatin and the centromere. Using this method, the centromere can be localized on the linkage map. Utilizing probes labeled with a variety of haptens, the utility of tetrad-FISH analysis would be enhanced. Signal disposition using multi-color FISH for each target sequence can reflect the positions of recombination. This method may also be applicable to animal models by observing signals in polar bodies. Ref., Kagawa N., Nagaki, K., Tsujimoto, H. (2002) Tetrad- FISH analysis reveals recombination suppression by interstitial heterochromatin sequences in rye, Secale cereale. Mol. Gen. Genet. 267:l0-15. (Hisashi Tsujimoto: Laboratory of Plant Genetics and Breeding Sciences. Faculty of Agriculture, Tottori University, Tottori 680-8553, Japan). |
|