Multicellular Organization Laboratory


Ihara’s
research
Regulation of Asymmetric division by intrinsic and extrinsic factors
Asymmetric divisions are indispensable for generating different cell types during development because they produce daughter cells that differ in fate. In stem cell, asymmetric division generates daughter cells that are committed to differentiation, while others maintain original stem cell fate. These different cell fate could be acquired either by unequal segregation of cell fate determinants or because of differential influences from surroundings, and these modes have defined either intrinsic or extrinsic mechanisms. The intrinsic regulation of asymmetric cell divisions are well known that division of Drosophila neuronblasts and the zygote of C.elegans are regulated by PAR proteins that are responsible for partitioning these determinants. The extrinsic mechanisms involve cell-cell communication and secretion signaling such as Notch and Wnt signaling.
Experimental model of asymmetric division regulated by Wnt signaling
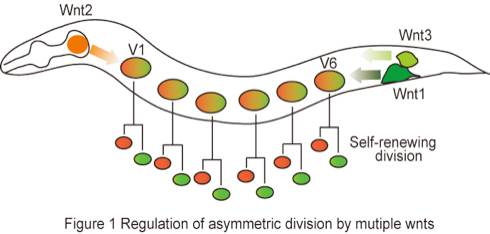
Although asymmetric division is fundamental process during development, most asymmetric division is experimentally inaccessible in mammal due to difficulties to analyze cell lineage in vivo, as they occur in complex and dynamic environments. Therefore, it is useful to study the asymmetric division in invertebrate model systems, especially Drosophila and C.elegans which exhibit similar properties of asymmetric division. By secretion of Wnts proteins, distinct cell populations could affect the regulation of gene expression in cells located at distant sites. In C. elegans, asymmetric division of most somatic cell occurring along the body axis (anterior-posterior axis) is regulated by a Wnt signaling. Wnt proteins are a family of extracellular ligands that are well evolutionarily conserved from worm to mammal. It is known that Wnts are modified by lipid and N-glycosylation. Although Wnt proteins were predicted to be soluble based on amino acid sequence, they are highly hydrophobic due to adding the lipid and stick tightly to cell membrane and extracellular matrix. These researches clearly indicate that post-translational modification alters the quality of Wnt proteins. In the past few years, rapid progress has been made in elucidating the mechanisms underlying Wnt signaling, but several outstanding questions remain to be addressed, in particular the relationships between the changed quality of Wnts by post-translational modification and extracellular matrix. To address such biological question, We will adopt six epithelial stem cells called seam cells (V1-V6) that repeat self-renewing divisions in C. elegans, as an excellent experimental model (Figure 1). Seam cells are hypodermal (epidermis) cells and asymmetric division is controlled by multiple Wnts ligands (H. Sawa: personal communication). Therefore, the seam cells are good model to elucidate Wnt properties in asymmetric division.
Aim1: Determine the post-translational modification of Wnts proteins in C.elegans
In C.elegans, post-translational modifications of Wnts proteins have not been reported on molecular levels. However, considering following facts, Wnts may be modified with lipidation and glycosylation in C.elegans. First, high conservation of the cysteine involved suggests that palmitoylation is common feature of all Wnts in C.elegans. Second, Porcupine (mom-1), which is homology to a family of o-acyl transferases, is conserved in C.elegans and may be involved in lipid modification. Third, potential N-glycsylation sites could be observed in Wnts protein, except for lin-44 (NetNGlyc 1.0 algorithm). From these results, We assure that dynamics of Wnts might be regulated by post-translational modification in C.elegans. We will identify post-translational modification of Wnts protein by biochemical method and analyze dynamics of Wnts protein in vivo. These results will be help to our understanding of Wnts function for asymmetric division of seam cell.

Copyright (C) 2010 NIG Multicellular organization lab. All Rights Reserved.


