Multicellular Organization Laboratory

Cell A







Specific characters of a variety of cell types are controlled by specific transcription factors. However, a single transcription factor often regulates multiple cell fates.


Cell fate determination
Most cells in C. elegans are produced by asymmetric cell divisions that are regulated by the Wnt/β-catenin asymmetry pathway. Therefore, POP-1/TCF that functions downstream of the Wnt pathway appears to be involved in determination of most cell fates. How does a single transcription factor regulate a variety of cell fates?


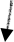



In the case of the T cell in the tail of the animals, we showed that specific cell fates are determined by the coordination between POP-1 and a Hox protein that regulate positional identity. Therefore, coordination with other transcription factors are important for specificity of POP-1 function.
In addition, we have recently reported that an acetylated-histone binding protein BET-1 is required for the maintenance of various cell fates. This suggests importance of epigenetic regulation for cell fate determination.
We will further study how POP-1 regulates various cell fates, focusing on the epigenetic regulation.


POP-1

NOB-1
psa-3 gene

POP-1


POP-1





POP-1







Cell A
Cell B
A model: POP-1 binding is determined by chromatin structure


ON
OFF


Transcription ON

Cell B


POP-1

NOB-1
psa-3 gene



Transcription OFF




Copyright (C) 2010 NIG Multicellular organization lab. All Rights Reserved.


